A new tool to ID spring dead spot
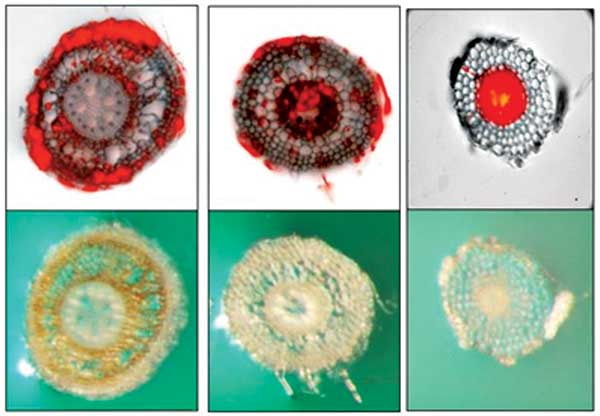
Colonization of a spring dead spot susceptible bermudagrass and cortical necrosis by Ophiosphaerella korrae (left), a tolerant bermudagrass (center) exhibiting vascular colonization by O. korrae and no necrosis and O. korrae colonization of a grass which does not produce disease (right). (Photo: Francisco Flores)
Bermudagrass is widely used as a turfgrass in warm regions throughout the world. Spring dead spot (SDS) is one of the most damaging bermudagrass diseases where cold temperatures cause winter dormancy. The causal pathogen is three fungal species of Ophiosphaerella, including O. herpotricha, O. korrae and O. narmari.
During cool temperatures, the fungi colonize the roots, stolons and rhizomes of the grass. The colonized stolons and rhizomes do not survive winter dormancy. The SDS symptoms appear as dead plants in circular patches anywhere from a few inches to 3 feet in diameter in the spring.
Weed establishment can be a problem in the dead areas and requires additional herbicide applications to reduce weed competition during bermudagrass regrowth.
Research shows that O. herpotricha is the most aggressive, followed by O. korrae and O. narmari. Smaller SDS patches may recover in the spring in a month, but large patches may require the entire growing season.
It is difficult to determine the Ophiosphaerella species because they seldom produce spore fruiting bodies. A correct diagnosis is needed to help turfgrass managers deal with the aggressiveness of the different species.
Researchers from Ecuador and Oklahoma State University developed a molecular assay to identify the three fungal species in the genus accurately. The team designed three pairs of DNA primers to identify fungal isolates and detect the pathogen in infected roots to achieve a species-specific diagnosis.
Results show the molecular technique can produce a unique DNA tag for each of the three SDS-associated Ophiosphaerella spp. The new molecular test does not detect the DNA of close relatives to Ophiosphaerella and common bermudagrass pathogens. This sensitive and specific molecular technique will correctly identify SDS-associated Ophiosphaerella spp. from field-collected roots, making this method a useful tool for timely diagnosis.
Reference
Martinez, J. Francisco Ituralde, Francisco J. Flores, Alma R. Koch, Carla D. Garzon and Nathan R. Walker. 2019. Multiplex End-Point PCR for the Detection of Three Species of Ophiosphaerella Causing Spring Dead Spot of Bermudagrass. Plant Disease 103(8):2010-2014. https://doi.org/10.1094/PDIS-10-18-1727-RE